> About us > Technologies > Next-Generation Sequencing
Next-Generation Sequencing, a high resolution, sensitive and cost-effective solution method for multiple gene analysis
Next-Generation Sequencing (NGS) offers a hypothesis-free approach that does not require prior knowledge of sequence information. NGS makes the simultaneous analysis of many genes possible and cost-effective. It is ideal for key stages in the workflow such as the acquisition of a cell line, banking and/or end of workflow.
Principles of Next-Generation Sequencing
Next-Generation Sequencing (NGS) technology has made whole genome sequencing (WGS) possible at an individual level. It allows the simultaneous analysis of many genes, which helps researchers understand the complexity of mutations in hPSCs.
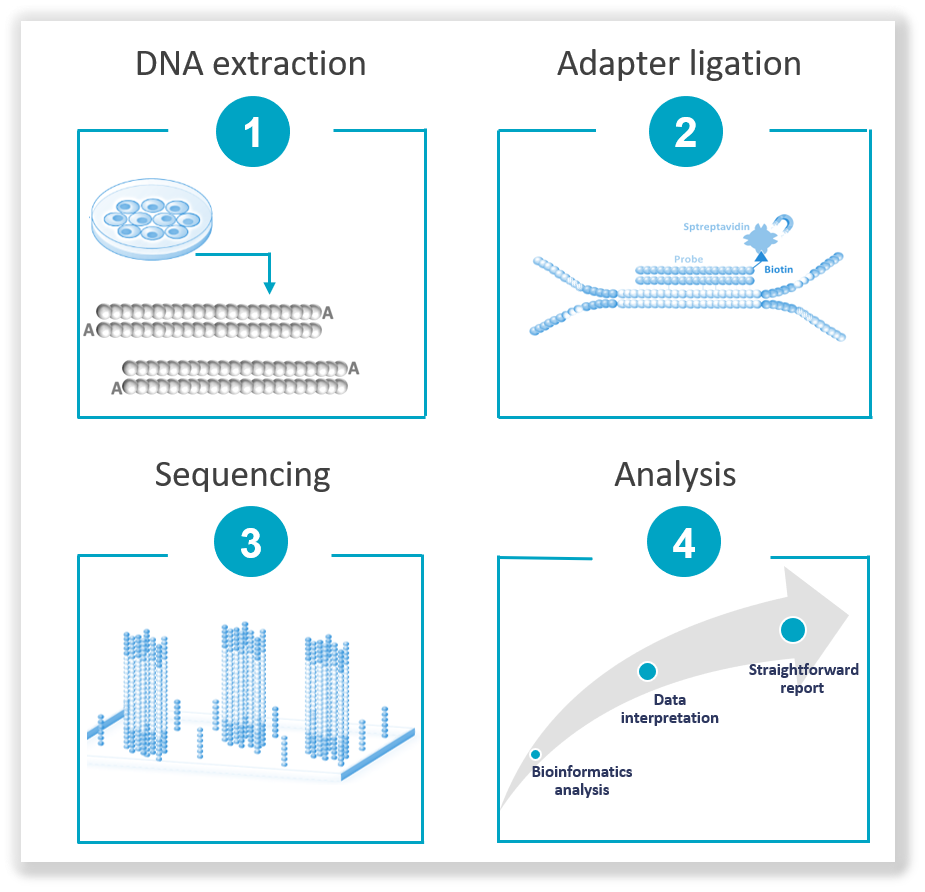
The NGS workflow typically involves several key steps, including sample preparation, sequencing library construction, sequencing and data analysis. During sample preparation, the DNA is extracted from the biological sample then fragmented into smaller pieces. These fragments are converted into a sequencing library by adding adapters and undergoing amplification. Then, the prepared library is loaded into a high-throughput sequencing platform, where millions of DNA fragments are simultaneously sequenced. The sequencing data generated is then processed and analyzed using bioinformatic tools to align the reads, identify genetic variants and interpret the results. The NGS workflow has significantly accelerated genomic research, enabled personalized medicine and contributed to our understanding of complex genetic diseases.
Targeted NGS panels are increasingly used to assess the value of gene mutations for clinical diagnostic purposes.
Capture sequencing versus amplicon
- The capture technique involves the use of specific probes or baits to capture the target sequences from the DNA sample. Then, the captured DNA fragments are sequenced using NGS technology. Capture sequencing allows for the sequencing of multiple genes simultaneously. This technique is well suited for sequencing a large number of genes or exons.
- The amplicon technique, on the other hand, uses PCR (polymerase chain reaction) to amplify the target regions of interest from the DNA sample. The PCR primers are designed to flank the target regions, and the amplified fragments are sequenced using NGS technology. Amplicon sequencing is typically employed for targeted sequencing of smaller regions, or a small panel of genes >500 kb. It can be challenging to design PCR primers that can efficiently amplify all regions of interest. In addition, the complexity of the PCR reaction can increase with the number of regions being targeted, which can lead to issues such as primer-dimer formation and non-specific amplifications.
For assay development, amplicon-based methods have been preferentially used for their shorter preparation time and small DNA input amounts. However, capture sequencing has emerged as an alternative approach because of its higher testing accuracy and reduced susceptibility to artifacts.
Comparison with other similar methods
- NGS versus Sanger:
Sanger sequencing allows the analysis of a single, distinct sequence in each sequencing reaction. In contrast, NGS enables the simultaneous sequencing of thousands of reactions from multiple samples within a single reaction.
Consequently, NGS enables cost-effective screening of a large number of samples and the detection of multiple variants across targeted regions of the genome. This approach would be both costly and time-consuming if performed by Sanger sequencing.
- Targeted NGS versus WGS:
Targeted sequencing offers several advantages over Whole Genome Sequencing (WGS). It is a cost-effective approach that focuses on specific regions of interest, resulting in reduced data generation, storage and analysis costs. Targeted sequencing achieves a higher depth of coverage in the targeted regions, improving sensitivity for variant detection. It is particularly useful for studying traits associated with specific genomic regions. The smaller data volume simplifies data management and analysis, enabling efficient variant calling and annotation. Targeted sequencing is scalable and suitable for large studies, providing a focused and cost-effective approach for investigating specific genomic regions.
Main advantages of Next-Generation Sequencing
- Empowers deep sequencing at high coverage levels for rare variant identification such as SNVs and Indels
- Drastically reduces sequencing costs and data analysis burdens compared with WGS
- Significantly reduces turnaround time compared to broader approaches
- Achieves lower limit of detection for enhanced sensitivity in variant identification
- Amplifies capacity with sample multiplexing, enabling efficient analysis of multiple samples
- Facilitates simultaneous sequencing of hundreds to thousands of genes or gene regions, expanding research possibilities and accelerating discoveries.

