> About us > Technologies > Digital PCR, high precision and speed
Digital PCR: high precision and speed.
Digital PCR (dPCR) is the most accurate PCR method for assessing the genetic integrity of stem cells through the detection of Copy Number Variants (CNVs).
Why?
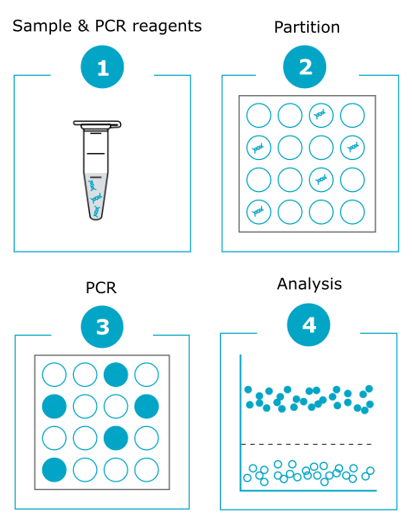
Using specific primer sets and fluorescent probes, dPCR amplifies targeted genomic fragments in thousands of individual reactions, thus allowing nucleic acid quantification and detection with high precision.
The power of dPCR is based on the random distribution of the sample and assay mixture into a very large number of distinct sub-reactions.
This distribution allows for multiple simultaneous endpoint PCR amplifications through the measurement and counting of positive (i.e. containing the amplified target) and negative (i.e. without the amplified target) partitions. The term ‘digital’ defines this binary result counting method. Positive reactions, which contain at least one copy of the target DNA fragment, are identified by their fluorescence signal. Comparison of the number of positive and negative reactions enables target quantification using the Poisson distribution method. Then, the absolute number of molecules present in the sample is calculated in comparison with the results obtained for a reference gene.
These features make dPCR an ideal tool for CNV detection with high sensitivity and precision.
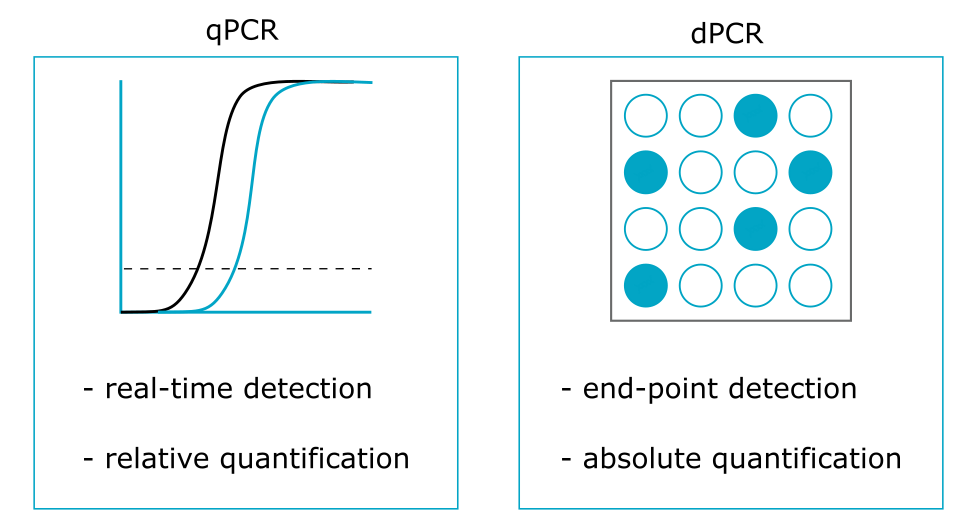
Comparison with quantitative PCR
Although it uses the same amplification reagents as dPCR, quantitative PCR (qPCR) is based on the analysis of a single homogeneous reaction mixture. Therefore, the number of template DNA copies is measured by real-time monitoring of the amplification at the exponential phase of the reaction, providing only a relative CNV quantification.
Main advantages of dPCR
The main advantages of dPCR compared with conventional qPCR methods are:
- Absolute quantification of nucleic acids without relying on external references (e.g. standard calibration curves);
- Robustness in terms of variations (thus, dramatic reduction of inter- and intra-assay variability) and tolerance of PCR inhibitors;
- High precision of CNV detection thanks to reaction partitioning (i.e. small fold differences can be revealed and this can be useful for the detection of low-frequency mosaic abnormalities).
For those who don’t have a dPCR machine
For those who have access to a dPCR machine