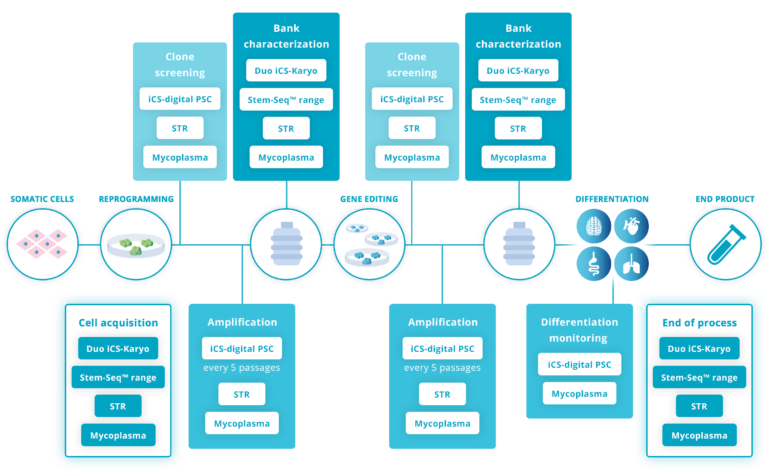
The high level of professionalism we find with our biotech clients working on human pluripotent stem cells (hPSCs) generally translates into well-established, robust quality controls.
They are aware of the propensity of hPSCs to develop abnormalities in culture (primarily CNVs), so they do everything they can to lower the chances of these genomic defects occurring.
To achieve this, they have put in place a series of methods, or “best practices”, that we want to share with you in this article.
1. High-resolution assays are necessary to ensure a stable hPSC culture
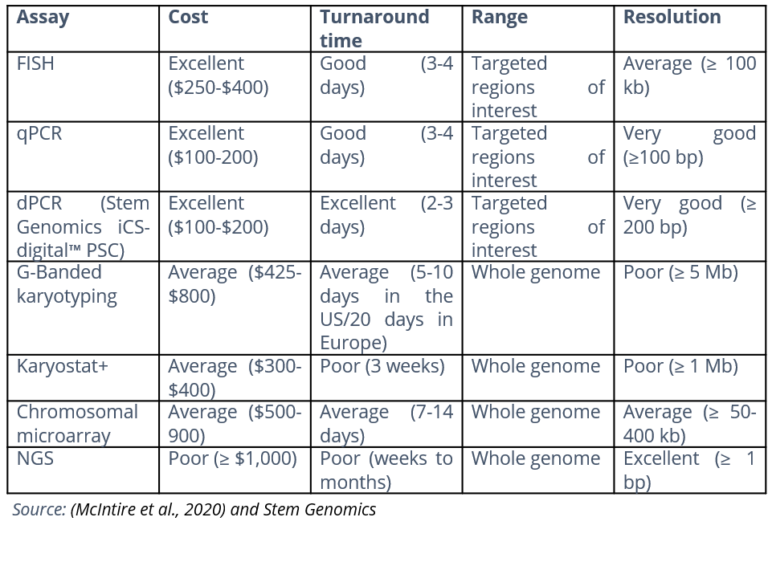
When it comes to QC for genomic stability of hPSCs, most publications mention G-Banding karyotyping. This will undoubtedly give you a very exhaustive numerical and structural assessment of your cells. However, it will present major drawbacks for hPSC lines/clones. With a resolution of 5 Mb, it will miss some of the most recurrent genomic alterations you can find in hPSCs, including the gain of chromosome 20q.11.21. This is the most frequent defect found in hPSCs, occurring in more than 20% of lines, and most of the time with no overt karyotyping changes (Amps et al. 2011).
The same can be said of assays like the Karyostat. They have better resolution than G-Banding (≥1 Mb) but can still miss recurrent abnormalities, particularly at early passages. Read Use Cases With Improved Resolution Leveraging dPCR Analysis by our client Synthego to see the performance of a high resolution digital-PCR based assay compared with Karyostat when it comes to the early detection of abnormalities in hPSC cell lines.
2. Fast turnaround is critical
Waiting for results to come in has a significant impact on the quality of your workflow and the confidence you can have in your cells, particularly in long-term cultures.
Therefore, choosing a test that can give you a first level of genomic stability information quickly and early in the process can make a huge difference.
Our client Cellected used to wait weeks for their G-Banding results to come back. In their case, this meant that they had already banked their cell lines and clones. When they started using our iCS-digital™ PSC test, they received results in days! This has enabled them to keep a closer eye on their cultures for the appearance of any sub-clones with copy number changes. They can now decide which cell line/clone is more stable and suitable for banking. See how they make the most of our dPCR assay.
For others like our client denovoMATRIX, adding our simple and cost effective iCS-digital™ PSC test to their process meant they could test their cells routinely every 10 passages. They have gained a higher degree of confidence by adding this simple test to their workflow. They admit that alternative methods would have been more painful. Read more about their experience.
3. Add some interim steps to your QC
Testing for genomic stability at the beginning and at the end of an hPSC culture is not sufficient. Best practice advises testing regularly during the maintenance and amplification stage, ideally every 5 passages (Assou et al., 2020; McIntire et al., 2020; Pamies et al., 2017).
Other key stages are:
– The acquisition of a new line: Read how our client DiNAQOR picked up a 20q abnormality on a recently purchased cell line. This can happen with very reputable providers.
– After gene editing: this step creates important stress on the cells, making CNVs more likely to appear. Read how CRISPR/Cas9 exposes your cells to genomic instability.
– At the bank characterization stage: Read how this made a big difference for Cellected
Using our digital PCR-based assay iCS-digital™ PSC in addition to other tests like G-Banding can massively increase confidence in the quality of your cells in long-term culture. Our client GoLiver explains how it has also rewarded them with a highly scalable, efficient and very cost-effective differentiation process.
digital PCR is a reliable tool when it comes to the detection of true positives. Our iCS-digital™ PSC false-positive rate is 0.098%, leaving very little room for error. It is well below the ≤5% usually expected for digital PCR-based assays.
4. Select the right mix of assays
How can you achieve a satisfactory test frequency without blowing your budget?
Your budget can only stretch so far, and achieving the most comprehensive quality control process is also about getting the right mix of assays.
We have put together this table that summarizes the key features of the most common genomic assay technologies.
For optimum, cost-effective in-process testing, we recommend associating the exhaustivity and sensitivity of G-Banding karyotype with a high-resolution digital-based PCR assay such as iCS-digital™ PSC.
To sum up, in order to complement your hPSC genomic quality control process, the main aspects to consider are:
– Performance: to detect sub-karyotypic abnormalities
– Speed: to enable the recommended level of routine screening of hPSCs within your time constraints
– Cost: so you can afford a routine testing strategy within your budget.
In our experience, our digital PCR-based assay ticks all the boxes and has proven to be easy to introduce into comprehensive, existing workflows that our biotech clients already had in place, adding significant quality to the end product.
Want to find out how this could be implemented in your own organization? Let’s talk!
Assou, S., Girault, N., Plinet, M., Bouckenheimer, J., Sansac, C., Combe, M., Mianné, J., Bourguignon, C., Fieldes, M., Ahmed, E., Commes, T., Boureux, A., Lemaître, J. M., & de Vos, J. (2020). Recurrent Genetic Abnormalities in Human Pluripotent Stem Cells: Definition and Routine Detection in Culture Supernatant by Targeted Droplet Digital PCR. Stem Cell Reports, 14(1), 1–8. https://doi.org/10.1016/j.stemcr.2019.12.004
McIntire, E., Taapken, S., Leonhard, K., & Larson, A. L. (2020). Genomic Stability Testing of Pluripotent Stem Cells. Current Protocols in Stem Cell Biology, 52(1). https://doi.org/10.1002/cpsc.107
Pamies, D., Bal-Price, A., Simeonov, A., Tagle, D., Allen, D., Gerhold, D., Yin, D., Pistollato, F., Inutsuka, T., Sullivan, K., Stacey, G., Salem, H., Leist, M., Daneshian, M., Vemuri, M. C., Mcfarland, R., Coecke, S., Fitzpatrick, S. C., Lakshmipathy, U., … Hartung, T. (2017). Good cell culture practice for stem cells & stem-cell-derived models. Altex, 34(1), 95–132. https://doi.org/10.14573/altex.1607121