The topic of quality control is growing in importance as the field of hPSC-based therapy becomes more mature. Research scientists that once only conducted basic checks for genomic stability are now going for more sophisticated assays and increased frequency of checks, often combing several methods. The aim is to gain reassurance concerning the quality of their final product. In line with the field’s evolving needs, Stem Genomics have worked closely with their clients to develop a range of complementary tests that can meet the QC requirements of an RUO environment. In this article, we tell you what to look for, how often you should test and which methods are suitable to achieve the desired genomic integrity of your cell line.
Which genomic variant should you be looking for?
The most frequent genomic abnormalities encountered in PSC are essentially copy number variations (CNVs) and single nucleotide variants (SNVs). CNVs reflect a selection pressure that may favor hPSC proliferation and survival, or reduce their differentiation capacities (Assou et al., 2018). As for SNVs, they can disrupt differentiation capacities, gradually take over the culture and increase the risk of cancer after transplants in patients. They can go unnoticed without a suitable assay. (Merkle et al., 2022).
The analysis of hPSC chromosomal abnormality would not be complete without an exhaustive structural and numerical variant analysis. This enables the identification of potential balanced and unbalanced translocations, aneuploidy, inversions and duplications/deletions, which are regularly cited in publications such as Halliwell et al. 2020.
At which stages should you be testing for genetic variants and why?
In the schematic figure below, you can see at a glance the key stages during which Stem Genomics recommend testing human Pluripotent Stem Cells (hPSC) in culture for genetic variants. Beyond avoiding the deleterious effects these defects can have on your final product, the aim is also to avoid the unnecessary waste of resources and time on potentially abnormal cell lines.
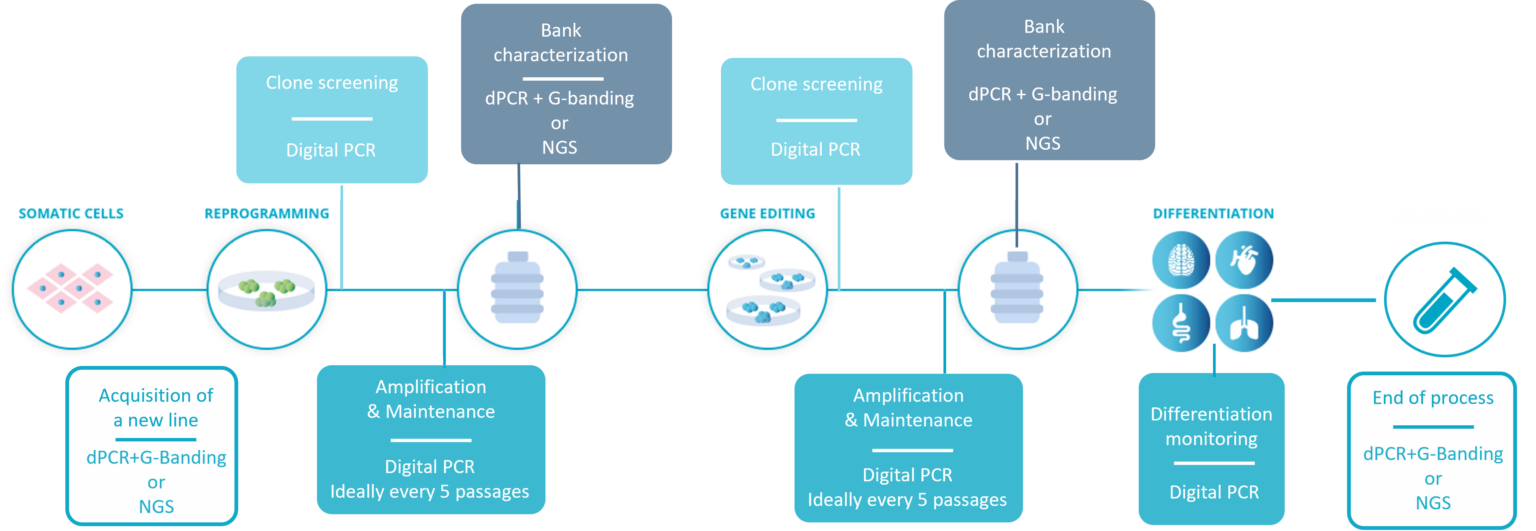
The workflow for hPSC genetic integrity quality control recommended by Stem Genomics.
- Acquisition of a new line: before starting any work, it is critical to ensure the genomic stability of the initial material. Most purchased lines will come with some kind of genomic stability tests, but we have encountered several instances of small size abnormalities not detected by G-Banding karyotype. Therefore, we recommend coupling G-Banding with digital PCR, or if your budget allows, running NGS testing.
- Reprogramming and gene editing: these procedures could favor the generation and selection of genomic aberrations in PSCs. A quick screening of clones and colonies is possible using digital PCR and is essential before any subsequent use.
- In-process control during cell amplification & maintenance: this constitutes another stressing factor likely to generate genetic defects in hPSCs. Considering the speed at which a recurrent abnormality can take over the culture, Stem Genomics and others (McIntire et al., 2020) recommend testing PSCs at least every 5-10 passages. This is realistically only feasible using digital PCR, as its multiplexing capabilities allow a high scope of detection with a fast turnaround and affordable cost.
- Pre-banking characterization: genomic stability is one of the critical features required for the complete characterization of a cell line before banking. For this stage, we offer two various methods that can be applied, and the scientist’s selection is often budget-dependent.
- Differentiation monitoring: we recommend continuing to check for genomic abnormalities until your cells have achieved full differentiation, and to do so when you change media. Once again, the speed of digital PCR makes it ideal for that stage.
- End of process: a final check will be requested before publication, but also before moving to the clinical stage. If you performed an NGS test at the start of the process, it is good practice to perform it again at the end of the process and compare the results. An alternative would be a G-Banding karyotype coupled with digital PCR, as it offers the usual structural rearrangement analysis combined with digital PCR’s resolution, which will pick up sub-karyotyping abnormalities such as 20q. This constitutes a suitable testing strategy at the end of an RUO process.
The methods we have selected for you:
Stem Genomics recommend using the following 3 methods for detection of genetically variant cells as approved by the ISSCR Standards:
1. Karyotyping: this is the standard technique for genome stability determination. It provides important information about numerical and structural alterations, and should be performed at least at the beginning and at the end of a project. This characterization method confirms with 98% probability that the tested cells are karyotypically normal. However, while useful, it cannot be regularly performed, and it is not sensitive enough to detect small genetic alterations (Baghbaderani et al., 2016). For a solution that addresses this issue, discover the Duo iCS-Karyo service designed for a comprehensive check on hPSC genomic stability.
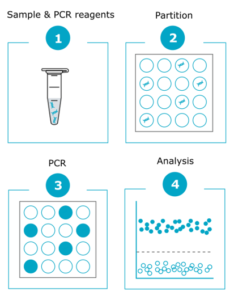
2. Digital PCR: Digital PCR offers higher resolution than karyotyping, and can detect abnormalities that cannot be identified by karyotyping, including chromosome 20q.11.21 amplifications that are the most common genetic abnormalities found in PSCs. Digital PCR amplifies targeted genomic fragments in thousands of individual reactions, thus allowing nucleic acid quantification and detection with high precision. Additionally, digital PCR is a fast method (results can be obtained within one day) and reasonably priced, more suitable for in-process testing as well as for screening a large number of clones after reprogramming or gene editing. If you’re looking for an off-the-shelf solution, discover our iCS-digital service, ideal for routine checks.
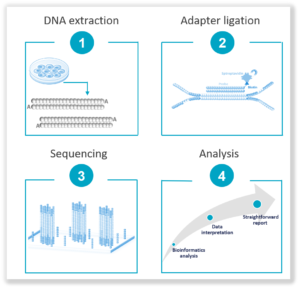
3. Next Generation Sequencing (NGS): It offers a hypothesis-free approach that does not require prior knowledge of sequence information. Highly sensitive with a low limit of detection, it empowers deep sequencing of high-coverage levels for rare variant identification such as SNVs and Indels and small size CNVs. It is ideal for key stages in the workflow such as the acquisition of a cell line, banking and/or end of workflow. Find out more about our NGS-based solution, the Stem-Seq Plus, a 2-in-1 solution for CNV and SNV detection, with interpreted results.
Assou, S., Bouckenheimer, J., & De Vos, J. (2018). Concise Review: Assessing the Genome Integrity of Human Induced Pluripotent Stem Cells: What Quality Control Metrics? Stem Cells, 36(6), 814–821. https://doi.org/10.1002/stem.2797
Merkle, F. T., Ghosh, S., Genovese, G., Handsaker, R. E., Kashin, S., Meyer, D., Karczewski, K. J., O’Dushlaine, C., Pato, C., Pato, M., MacArthur, D. G., McCarroll, S. A., & Eggan, K. (2022). Whole-genome analysis of human embryonic stem cells enables rational line selection based on genetic variation. Cell Stem Cell, 29(3), 472-486.e7. https://doi.org/10.1016/j.stem.2022.01.011
Halliwell, J., Barbaric, I. & Andrews, P.W. Acquired genetic changes in human pluripotent stem cells: origins and consequences. Nat Rev Mol Cell Biol 21, 715–728 (2020). https://doi.org/10.1038/s41580-020-00292-z
Baghbaderani, B. A., Syama, A., Sivapatham, R., Pei, Y., Mukherjee, O., Fellner, T., Zeng, X., & Rao, M. S. (2016). Detailed Characterization of Human Induced Pluripotent Stem Cells Manufactured for Therapeutic Applications. Stem Cell Reviews and Reports, 12(4), 394–420. https://doi.org/10.1007/s12015-016-9662-8
McIntire, E., Taapken, S., Leonhard, K., & Larson, A. L. (2020). Genomic Stability Testing of Pluripotent Stem Cells. Current Protocols in Stem Cell Biology, 52(1). https://doi.org/10.1002/cpsc.107

