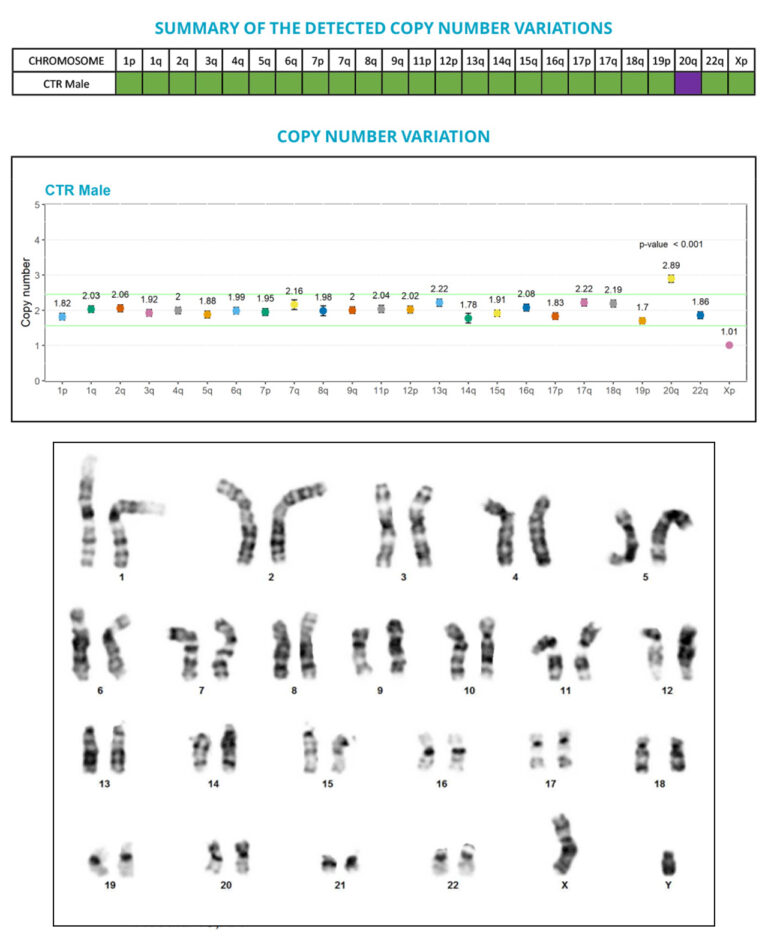
“Cell preparation is hell!”, “It didn’t detect a 20q abnormality”, “It takes months to get our results back…”
Sounds familiar?
These are some of the complaints that we hear from many research scientists working on PSCs when they relate their experience of G-banding karyotyping.
It is true that the test is demanding. It requires you to prepare a robustly dividing cell culture in order for the assay to be successful. And that is the step before the chromosome analysis. It also requires specialized training. As a result, it is challenging for most researchers to conduct it in-house.
Could you do without?
The G-banding karyotype is firmly established within the stem cell community as the “gold standard” in terms of genetic stability measurement. It has been around for decades, which also plays in its favor as it has enabled consistent record keeping through standardized nomenclature (Liehr, 2021).
But is it here to stay?
In their paper, McIntire et al. describe karyotyping as a well-established assay, historically used for PSC genomic testing, while NGS and PCR represent up-and-coming alternatives (NGS for high-throughput characterization, PCR for in-process).
We could not agree more. It’s safe to add that the right strategy for you will very much depend on the desired level of precision of your quality control procedure, the allocated budget and how long you can afford to wait for your final analysis.
As a result, we observe that most of our clients still turn to karyotyping for characterization at key stages in their workflow, typically when acquiring a new line and at the bank characterization stage. After all, it is currently one of the most robust assays out there.
However, one limitation of this assay for PSCs is that it regularly misses some recurrent abnormalities due to its low resolution (> 5 Mb). And that’s where we come in!
The key: add a high-resolution test to your karyotyping!
We have developed a combined solution that we are confident will bring the reassurance you need for the genomic integrity of your cells.
We looked for a complementary solution with high-resolution testing and selected dPCR. As you probably know, dPCR is a more advanced PCR technology than qPCR and therefore provides increased sensitivity. More on dPCR.
Thanks to the performance of the digital PCR technology, we developed a test that allows the sensitive and precise quantification of copy number variations (CNVs), and the detection of anomalies that are below the G-banding method resolution, especially the sub-karyotypic 20q11.21 amplicon.
Our expertise in PSCs and digital PCR has enabled us to bring you a tailored solution for your specific needs, which we call Duo iCS-Karyo. Capitalizing on the strengths of both G-banding karyotyping and the iCS-digital PSC-24 probes test, it provides you with the comprehensive genomic analysis that karyotyping can give you with the targeted resolution from iCS-digital™ PSC technology. It’s designed to help you characterize your cell lines at the acquisition and banking stages.
Our promise to you, in a nutshell:
✅ We take care of cell preparation for you
✅ You get high-resolution detection of the most recurrent altered regions in PSCs
✅ Combined with a low-level mosaicism, genome-wide analysis
✅ All this delivered to your inbox in the form of a straightforward analysis with reasonable lead-times (up to a month)**
How does that sound? Contact us if you’d like to give it a try.
** Assuming the samples sent comply with the sample collection and shipment instructions. Please note that we will not be able to plan the work on your cells without advance notice. Contact us at least one week before sending your samples to avoid disappointment.
Liehr, T. (2021). International System for Human Cytogenetic or Cytogenomic Nomenclature (ISCN): Some Thoughts. In Cytogenetic and Genome Research (Vol. 161, Issue 5, pp. 223–224). S. Karger AG. https://doi.org/10.1159/000516654
McIntire, E., Taapken, S., Leonhard, K., & Larson, A. L. (2020). Genomic Stability Testing of Pluripotent Stem Cells. Current Protocols in Stem Cell Biology, 52(1). https://doi.org/10.1002/cpsc.107