Digital PCR, also known as dPCR, was first described by Sykes et al. in 1992 (Sykes et al., 1992) and although it has been around for a while, it is not as widely spread as qPCR worldwide. Qualified as a “robust technique”, those who have adopted it appreciate the precision and sensitivity it offers. Lately it has been experiencing accelerated adoption by laboratories in response to the rise in prevalence of infectious diseases, cancer and genetic disorders and the more frequent introduction of off-the-shelf products. Particularly relevant for certain fields, it has emerged as a reliable technique for quantifying target sequences and identifying rare variants. It was therefore a logical choice for Stem Genomics’ founders back in 2018.
How does it work?
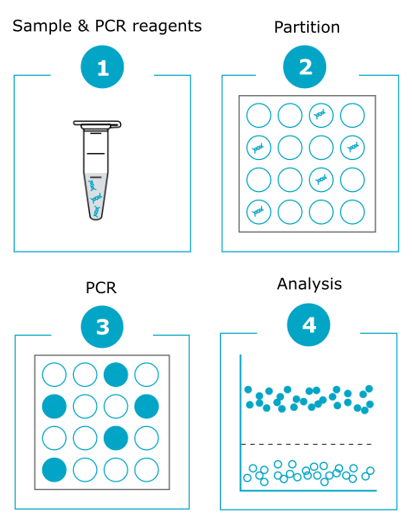
A single reaction takes place in thousands of nanoliter-sized compartments (ranging from 8,500 to 30,000 partitions depending on the platform used) (1). This distribution allows for multiple simultaneous endpoint PCR amplifications (2) through the measurement and counting of positive (i.e. containing the amplified target) and negative (i.e. without the amplified target) partitions. The term ‘digital’ refers to this binary result counting method (3). Positive reactions, which contain at least one copy of the target DNA fragment, are identified by their fluorescence signal. The comparison of the number of positive and negative reactions enables target quantification using the Poisson distribution method (4).
💡Other digital PCR methods such as chip-based digital PCR, using a plate partitioning method, are becoming increasingly popular. This is attributed to its high-performance in detecting a strain’s specificity with less variability compared to the current standard of plate count enumeration techniques and quantitative polymerase chain reaction (qPCR) (Source: Fortune Business insights).
Fields where dPCR is particularly relevant
Digital PCR is ideal for complex samples with a low concentration of target molecules, making it very popular in biopharma applications, oncology and organ transplantation fields.
Most digital PCR applications require exceptionally high sensitivity, specificity and discriminatory power. Such digital PCR applications include:
- Rare sequence detection
- Analysis of copy number variation (CNV)
- Quantification of rare mutations including in liquid biopsy testing
- Single-cell analysis
- Measurement of viral load
- Gene expression and miRNA analysis
- Pathogen detection
- Quantification of next-generation sequencing (NGS) libraries
- Detection of rare targets from environmental samples, including wastewater or sewage
🧫In the stem cell field, dPCR is recognized as the most accurate PCR method for assessing the genomic stability of stem cells (CNVs, SNVs…). Digital PCR can achieve a sensitivity of up to 0.1% (in certain situations even up to 0.001%), compared to 1% for the traditional PCR method (Lambrescu et al., 2022). Stem cell research scientists use it to detect genetic variants of various sizes, and in particular the infra-karyotypic mutations that G-Banding cannot detect. Thanks to the intrinsic properties of dPCR technology, high precision in detection and quantification can be achieved. It is also a technology of choice for detecting residual undifferentiated PSCs, a major safety issue for iPSC clinical applications due to the potential risk of PSC-derived tumor formation (Yasuda et al., 2024).
All in all, digital PCR is expected to grow in importance as precision medicine becomes more commonplace.
Why do Stem Genomics’ clients choose to add the iCS-digital™ range of tests to their workflow?
The speed, sensitivity and cost-efficiency of this technology give digital PCR an edge over other genomic stability testing techniques and methods.
- Speed
For many biotechs, producing fast is crucial to responding to their clients’ demands. Speed also makes it possible to test more often, increasing scientists’ confidence level in their final product.
The QC department at Cedars-Sinai uses the iCS-digital™ PSC assay to check genomic stability in-process. It is part of a comprehensive range of tests selected for their complementarity in achieving optimum quality control. They value the 1-day turnaround of the iCS-digital™ PSC assay (they use the in-house kit) that enables a quick go/no go decision before cell banking and manufacturing final cell products.
For EditCo, the iCS-digital™ PSC assay is the only technology compatible with the speed and scale that they need as part of their automated cell-editing workflow. “We evaluated a lot of karyotyping technologies and only Stem Genomics’ digital PCR-based assay enabled the scalability, sensitivity, repeatability and speed we needed to rapidly iterate and improve our processes.”
For GoLiver, the test has been a great help in detecting genomic abnormalities earlier in the process. According to Angélique Fourrier, R&D project manager at GoLiver, “since we started using the iCS-digital™ PSC for routine control, we have gained a massive degree of confidence in the quality of our cells in long-term culture, as we can quickly control their genome integrity, even on a weekly basis if we wanted”.
- Performance
Thanks to its high-sensitivity, digital PCR enables the detection of sub-karyotyping abnormalities smaller than 5-10 Mb (G-Banding resolution). This is particularly crucial regarding the gain of 20q.11.21 copy-number variant (CNV). The 20q.11.21 chromosomal amplification, commonly known as 20q, is detected in more than 20% of cultured human Pluripotent Stem Cells (hPSCs) worldwide (Avery et al., 2013; Halliwell et al., 2020) and accounts for 22.9% of the recurrent structural variants identified in these cell lines (Assou et al., 2020), making it the most common genomic abnormality in hPSCs. It is therefore critical to use an appropriate technique for detection at the required sensitivity level.
This is what Kurt Jacobs, a research scientist at DiNAQOR, experienced when using the iCS-digital™ PSC kit: “Stem Genomics’ assay has helped us strengthen our quality control at various stages of our workflow. For instance, it picked up the 20q amplicon that was present in some of our hiPSC lines.”
Renee Maas from The Regenerative Medicine Center in Utrecht reached the same conclusions. According to Renee, “since establishing a QC strategy involving iCS-digital™ PSC, we realized that the 20q appears very often. We really did not expect it to be present so frequently! This led us to implement iCS-digital™ PSC every 10 passages”.
- Cost
In 5 passages or less, the variants that appear can rapidly become predominant, compromising iPSC-based research work (Assou et al., 2020; McIntire et al., 2020; Pamies et al., 2017). It is therefore important to pick up abnormalities as early as possible in an iPSC workflow, and G-Banding (the traditional method for genomic stability testing in PSCs) is not designed to support that level of frequency in testing*. The iCS-digital™ assay range makes regular in-process testing possible every 5-10 passages thanks to its cost efficiency.
It also enables clone screening after gene editing without bumping up costs and while saving money down the line. The iCS-digital™ range provides first-level screening that is fast and cost effective before selecting the clones that will go through further genomic stability testing.
*We recommend integrating the iCS-digital™ PSC with G-Banding at specific stages of the workflow. Find out more here.
Latest update from digital PCR expert, Juline Vincent, R&D Project Manager at Stem Genomics
What led you to specialize in digital PCR?
After general university studies in biology, I was lucky to be exposed to several platforms using digital PCR. When I moved to Stem Genomics, a new field of application opened up for me with the analysis of copy number variations (CNV) in iPSCs. The latest advances in the digital PCR field allow Stem Genomics’ tests to be highly multiplexed. This is quite challenging to develop and optimize, but there are many advantages.
What are the technical advantages that dPCR offers stem cell scientists?
Several tens of thousands of partitions are generated, allowing robust statistical calculations to define a Copy Number Variation with high accuracy. At Stem Genomics, we look at the total number of partitions generated to ensure that CNV results are as accurate as possible. Thanks to the new digital PCR platforms and their multiple fluorescent channels, the multiplexing capability has increased. That’s why, today, Stem Genomics can offer a test with 28 probes for the same price as the previous 24 probes and the same amount of DNA. This allows our clients to save time and money.
What do you find to be the most exciting developments in the world of digital PCR lately?
When I began using digital PCR, there was only one manufacturer, but today, new players are emerging on the digital PCR market. They are well-known companies that have decided to launch their own digital platform. This shows the incredible rise and the promising future of this technology. All these new technologies use different types of suitable partitions (droplets or chip-based partitions) with a variety of samples and offer a large range of test throughputs. In other words, there’s now something for everyone in digital PCR. Manufacturers have also improved polymerase enzymes and added new fluorophores channels, which allow users to increase their multiplexing capabilities.
What are the next developments going to be for Stem Genomics’ clients?
The new challenge for Stem Genomics will be the analysis of pluripotency gene expression through the analysis of iPSC RNA. This means that the test won’t analyze CNV but rather a relative expression. We’re not giving up DNA, and hope to push back the limits of multiplexing with new tests and maybe new digital PCR platforms… stay tuned!
Discover Stem Genomics' range of digital PCR assays:
For those who don’t have a dPCR machine
For those who have access to a dPCR machine
Sykes, P. J., Brisco, M. J., Hughes, E., & Morley, A. A. (n.d.). Quantitation of targets for PCR by use of limiting dilution Minimal residual disease in leukemia View project Parthenolide and prostate cancer View project. https://www.researchgate.net/publication/21766890
Lambrescu, I., Popa, A., Manole, E., Ceafalan, L. C., & Gaina, G. (2022). Application of Droplet Digital PCR Technology in Muscular Dystrophies Research. In International Journal of Molecular Sciences (Vol. 23, Issue 9). MDPI. https://doi.org/10.3390/ijms23094802
Yasuda, S., Bando, K., Henry, M. P., Libertini, S., Watanabe, T., Bando, H., Chen, C., Fujimori, K., Harada, K., Kuroda, T., Lemmens, M., Marginean, D., Moss, D., Pereira Mouriès, L., Nicholas, N. S., Smart, M. J. K., Terai, O., & Sato, Y. (n.d.). Detection of residual pluripotent stem cells in cell therapy products utilizing droplet digital PCR: an international multisite evaluation study. https://doi.org/10.1093/stcltm/szae058/7730734