The many upcoming clinical trials using hPSCs and hPSC-derived cells will require more rigorous genomic testing to understand the cells’ mutational landscape and the potential risks when using them. In other words, selecting lines based on convenience or historical precedence rather than intrinsic genetic stability (Kobold et al., 2015) will no longer be acceptable for the stem cell community, even in the research stage.
Importance of identifying genetic variants associated with cancers in hPSC
Advanced testing technologies have assisted in the discovery of new genomic abnormalities. These abnormalities are primarily chromosomal defects and point mutations (SNVs) that can spontaneously appear in hPSC lines in prolonged cultures. Genetic variants associated with cancers are amongst the most frequently observed genomic defects. Without a suitable assay, they can go unnoticed and disrupt differentiation capacities, gradually take over the culture and increase the risk of cancer after transplants in patients (Merkle et al., 2022). This also applies to development studies and disease modeling where those abnormalities can compromise the end product quality. Therefore, it has become critical to regularly assess the genomic stability of hPSC lines with an efficient testing strategy.
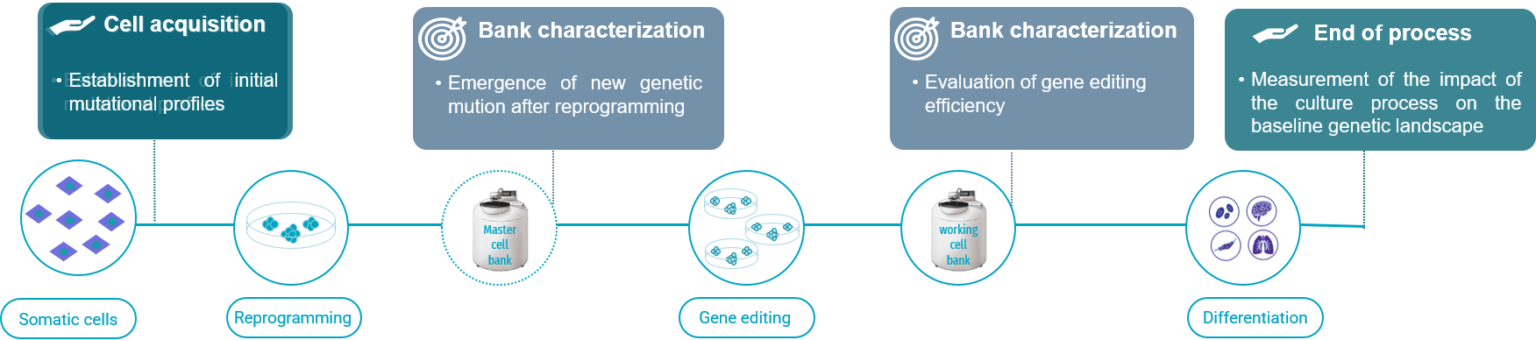
Recommended guidelines for a robust genomic stability hPSC workflow
Thanks to their high sensitivity, sequencing assays are the only suitable methodology to fully capture variation in the resolution of nucleotides in hPSCs (Lezmi & Benvenisty, 2021). This is why Stem Genomics recently completed its range with a powerful NGS-based assay designed to meet the evolving needs of stem cell research scientists.
Stem-Seq™ panel: a highly-sensitive NGS assay dedicated to hPSCs
Most NGS-based panel assays identifying oncogenic variants available on the market are designed for general clinical diagnostics. They are used by default by stem cell research scientists, but have some limitations:
- the genes selected are not always representative of the recurrent variants found in hPSC lines, such as TP53 (Lezmi & Benvenisty, 2021; Merkle et al., 2017) or BCOR (Rouhani et al., 2022) and/or the gene panels are too small to cater to the abundance of potential mutations in hPSCs.
- the result interpretation is not adapted for PSC research scientists, who can become lost in the vast amount of data produced, complicating the decision-making process.
Discover Stem Genomics’ Stem-Seq™ panel, an assay that precisely meets the growing needs of hPSC scientists for high-power detection assays.

